Cardiac Electrophysiology - I
In this blog I will show you can configure the DuneCopasi simulation environment to perform a very simple cardiac electrophysiology simulation. I will show you how you can simulate the Mitchell-Schaeffer cardiomyocyte electrophysiology model in a 2D environment using the DuneCopasi Docker container.
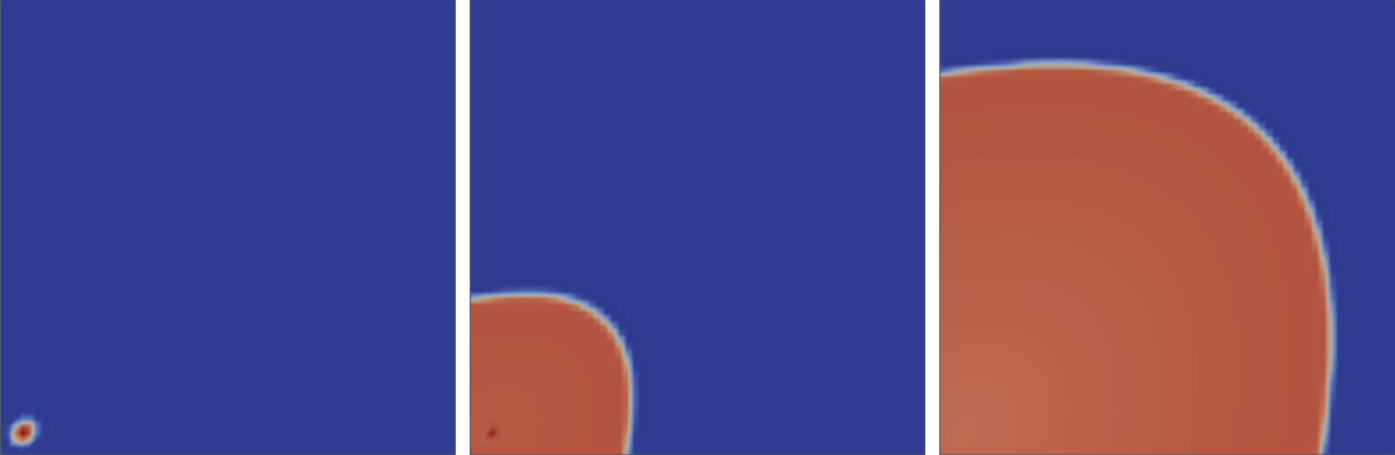
We will simulate the Mitchell-Schaefer model where we stimulate the bottom left corner using a Gaussian point source. This will result in the activation and repolarization of our 2D square grid. Lets get started.
The mathematical model of cardiac electrophysiology
The Mitchell-Schaeffer model of cell electrophysiology
Cardiomyocytes maintain an ion concentration gradient between the intracellular and extracellular space. This ion concentration gradient results in a potential across the cell membrane (i.e. the transmembrane potential). A typical resting membrane potential is around . A disturbance of this resting state can result in a cardiac action potential. This is a brief change in membrane potential of heart cells resulting in a typical transmembrane potential waveform. This typical waveform is determined by the dynamics of the ion channels in the cell membrane. In the very simple Mitchell-Schaeffer model the action potential is described by 2 ion channel currents. The governing equations are given by
where and are time-scale variables, and describe the respective inward and outward current, is the normalized transmembrane potential and is a gating variable. The dynamics of the gating variable are given by
where and are time-scale variables for the gating variable and defines the critical threshold for depolarization.
The bi-domain equation of cardiac electrophysiology
In the section above we described a model capturing the basic electrophysiology of cardiomyocytes. Now we want to extend this by coupling multiple cardiomyocytes yielding a spatial description of the cardiac electrophysiology. To obtain this we will depart from Ohm's law stating
In our specific case we have two compartments. We will use an intracellular and extracellular current density and potential. We obtain
Furthermore, we assume that any current leaving the intracellular space flows into the extracellular (and vice-versa) and that there is no charge accumulation. Therefore, the change in current density in the extracellular space has to be equal to the current flowing into the intracellular space. Using Gauss' law (i.e. the divergence theorem) this can be written as
The current flowing from the intracellular into the extracellular space is nothing else than the transmembrane current (per surface area). The conservation of current can be rewritten as
The transmembrane current can be modeled using the cable equation where we combine the ion channel current with a capacitative component
where is the transmembrane potential and is the surface density. Rewriting these equations (by eliminating ) yields the standard form of the bidomain equation of cardiac electrophysiology:
EP simulations using DuneCopasi
Now that we have discussed the mathematical problem we can tend our attention to implementing this so that we can simulate it using DuneCopasi. To do this we will write a configuration file named cardiac_ep_1.ini that will entail all the information that is needed for the DuneCopasi solver to construct the problem at hand.
We start by creating a working directory where we will save the simulation files.
~$ cd workdir
~/workdir$
Setting up the Mitchell-Schaefer model
Let us get started with creating the ini file for our electrophysiology simulation. Open your favorite text editor and you can start building the configuration file. First we will define our grid.
The grid properties
The grid is defined as follows:
[grid]
dimension = 2
extensions = 3 3
refinement_level = 8
This will create a square grid existing of 2 triangles with 3 by 3 extension. Thereafter the grid will be refined 8 times. The solver can also load Gmsh meshes but we will come to that at a later point. For the moment this will be all that we need to define our grid. The next step is the parser_context.
The parser context
In the parser context we will define the constants and functions that we will use in our model. This is easiest seen by looking at an example:
[parser_context]
tau_in.type = constant
tau_in.value = 0.1 # ms
tau_out.type = constant
tau_out.value = 1 # ms
tau_open.type = constant
tau_open.value = 150 # ms
tau_close.type = constant
tau_close.value = 120 # ms
gamma_i.type = constant
gamma_i.value = 3e-3 # 1/(\Omega*cm)
A_m.type = constant
A_m.value = 20e-1
C_m.type = constant
C_m.value = 1.0
u_0.type = constant
u_0.value = 0.13
gauss.type = function
gauss.expression = x, y, z: exp(-(x^2+y^2+z^2)/(4*0.0001)) / (4*3.14159265359*0.0001)/400
pulse.type = function
pulse.parser_type = ExprTk
pulse.expression = t, t0, dt: sqrt((t-t0)^2)< dt ? 1 : 0
To create a constant variable lets say tau_in we simply write tau_in.type = constant)). The value of the constant then assigned by writing tau_in.value = 0.1. In a similar fashion a function is defined. For our Gaussian function which we name gauss we simply write gauss.type = function. The function itself is then defined by typing gauss.expression = x,y,z : exp(... where the x,y,z before the colon denote the function arguments and the function itself is defined on the right of the colon. Note that standard mathematical functions such as exp, sin, etc. are available from the underlying parser allowing great freedom to define mathematical expressions. At this point we already have a grid and defined some constants and functions, now we will start defining the problem.
Defining the compartments
Although this example is fairly simple and we only need 1 compartment (which is the complete square), the concept to define multiple compartments is similar and therefore we will already discuss this briefly. Defining a compartment is similar to defining a defining a function. For our example we have one compartment which we will call the domain. To define this compartment type under the compartments section domain.type = expression. This indicates that we will provide an expression which will be evaluated for each entity and if this expression evaluates to be non-zero then it will be added to the compartment. Here we only have one compartment which is all of the space so we set this expression equal to the constant 1.
[compartments]
domain.type = expression
domain.expression = 1
The model properties
The first step in defining the model is to chose the parser that will be used to define the problem. Here we will go for ExprTk.
[model]
parser_type = ExprTk
Once that is selected we will start defining the first variable of our problem. In this case we will start with the transmembrane potential which we refer to as . To do this we will create a subsection model.scalar_field.V with the properties of our problem.
[model.scalar_field.V]
compartment = domain
storage.expression = 1
cross_diffusion.V.expression = gamma_i / (A_m * C_m)
cross_diffusion.phi.expression = gamma_i / (A_m * C_m)
reaction.expression = (z*V^2*(1-V)/tau_in - V/tau_out)/C_m + 0.85*pulse(time, 10, 20)*gauss(position_x - 0.15, position_y - 0.15, position_z)
reaction.jacobian.V.expression = (z*V*(2-3*V)/tau_in-1/tau_out)/C_m
reaction.jacobian.z.expression = ((1-V)*V^2/tau_in)/C_m
initial.expression = 0.01
The compartment property will define the compartment on which the scalar_field is defined. Here we only had one compartment domain that corresponds to the complete grid. Since the equation determining is time-dependent we have to store the previous time-step for use in the time-stepping scheme. We do this by setting storage.expression to 1. The following 2 lines define the cross diffusion of respectively and . Then we define the reaction term of the problem and the associated Jacobian entries of the reaction term. Note that we defined a function in the reaction term explicitly using the position and time to assemble the problem. At last we provide a function defining the initial state of the problem. In a similar fashion we define the extracellular potential field .
[model.scalar_field.phi]
compartment = domain
storage.expression = 0
cross_diffusion.V.expression = gamma_i / (A_m * C_m) # (gamma_i)/(A_m * C_m)
cross_diffusion.phi.expression = 1.25 * gamma_i / (A_m * C_m) # (gamma_i + gamma_e)/(A_m * C_m)
reaction.expression = 0
constrain.boundary.expression = ((position_x < 0.025) & (position_y < 0.025)) ? 0.0 : no_value
initial.expression = 0.00
The equation is not time-dependent so we set storage.expression to zero. Further, we add the cross diffusion components and set the reaction term to zero. Note that explicitly setting the reaction term to zero is not necessary as the solver sets all non-defined properties to zero. Thereafter, we add a constraint to a small boundary region in the bottom left corner. Note that adding this constraint is technically not necessary, but since the variable only appears as gradients the solution is only determined up to a constant. So by setting the bottom left corner to zero we consider the potentials to be with respect to the potential in the bottom left corner. At last we provide the initial value. Similarly we define the gating variables .
[model.scalar_field.z]
compartment = domain
storage.expression = 1
reaction.expression = (V <= u_0) ? ( (1 - z)/(tau_open) ): (-z/tau_close)
reaction.jacobian.z.expression = (V <= u_0) ? ( -1/(tau_open)) : (-1/tau_close)
initial.expression = 1
Since these equations has no diffusion components, we simply don't define them rendering the coefficients of the diffusion and cross-diffusion zero. By defining the problem as time-dependent with a reaction term and no diffusion this is in fact just a ordinary differential equation. So at this point we have defined the mathematical properties of the problem. The next step is to define the underlying time-step approach to solve these equations. This is done by setting the model.time_step_operator properties.
[model.time_step_operator]
type = ImplicitEuler
time_step_initial = 0.500
time_step_max = 0.500
time_end = 800
linear_solver.type = BiCGSTAB
For this problem we will use an implicit-Euler time-stepping scheme and then we define the initial and maximal time-step. The simulation will step from time-point time_start to time_end. In our case this will be from (default value) to . To solve the linear problem we will use the BiCGSTAB iterative solver. The last step is to write the solution to a file. This is done by initializing the writer.
[model.writer]
time_step = 1.000
vtk.path = 2D_Mitchell_Schaefer_model
At this point we have defined all the properties of the problem and we are ready to simulate the model! Let us save the configuration file to cardiac_ep_1.ini.
Click here to expand/collapse the complete configuration file
Running the Mitchell-Schaefer model
Running the code is the same as described in the Docker Documentation, simply call:
[2024-03-21 12:38:32.479] [info] Reading configuration file '~/workdir/cardiac_ep_1.ini'
[2024-03-21 12:38:32.479] [info] Starting dune-copasi (version: 2.0.1)
[2024-03-21 12:38:32.580] [info] Axis names are set to: ["x", "y", "z"]
...
...
[2024-03-21 12:38:32.723] [info] dune-copasi successfully finished :)
~/workdir$
The simulation will start and write the *.vtk files to the specified directory. You can easily inspect these files by opening them in Paraview.

At this point we have successfully simulated the activation of a 2D square grid using the Mitchell-Schaefer cell model. Although this simulation model is very simple it does already illustrate the capabilities of the DuneCopasi solver. In the next post we will dig deeper into the available options and explore true multi-compartmental problems.